Overview
Our focus on function makes us unique!
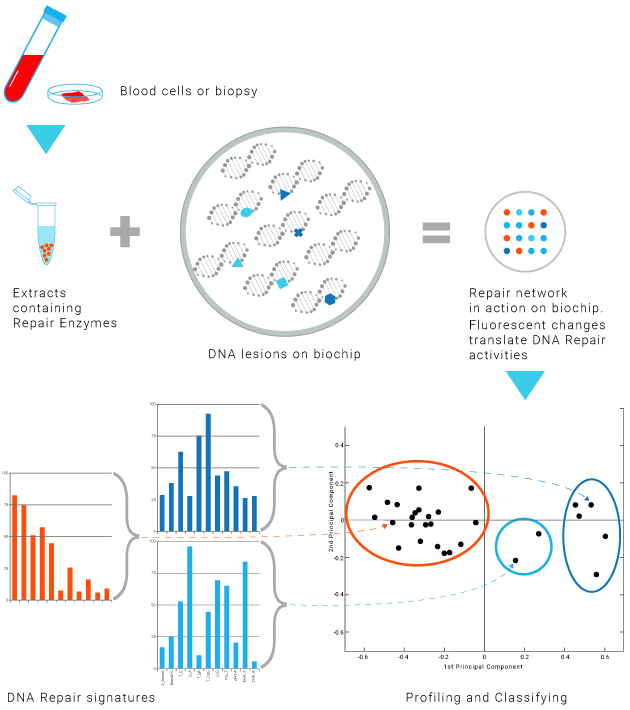
Innovative method
Tumor biopsy
Blood cells
DNA Repair enzymes contained in a biological sample in contact with series of lesion-containing DNA on a biochip.
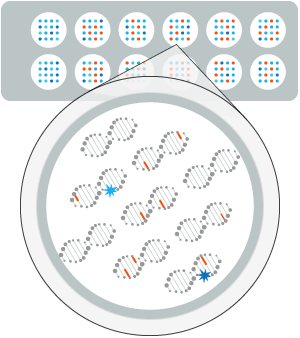
Fluorescence analysis reveals the repair reaction occurring at each lesion site. —

The efficiency and accuracy of the DNA Repair mechanisms is profiled for each patient.
This profiling provides insights into the patient' ability to respond to a treatment.
Focusing on function rather than on genes
We focus on functional profiling rather than on genomic profiling

In addition, the result takes all possible molecular regulation into account (epigenetic, transcription, pathway reactivation, etc).
DNA Repair mechanisms, DDR and cancer treatments, an integrated view
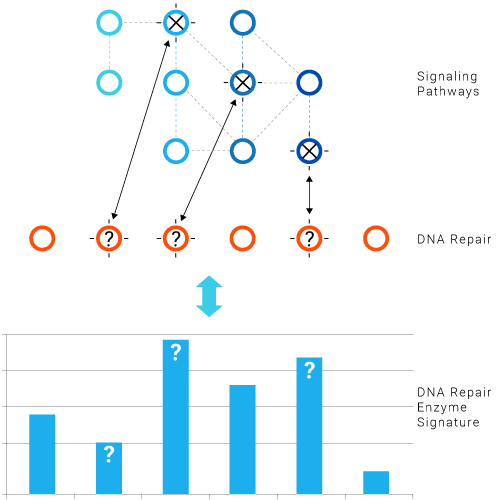
Major genes in these networks are frequently mutated in cancers.
- Mutations in the signaling cascade have functional consequences that affect DNA Repair and DDR in different ways.
- The DNA Repair Enzyme Signature reflects the impact of driver mutations in the signaling cascade.
- Driver mutations in the signaling cascade and mutations in DNA Repair or DDR genes determine the response to cytotoxic drugs, radiotherapy, targeted therapies and DNA Repair inhibitors.
- Pathway redundancy makes the system very complex and the functional approach highly relevant.
DNA Repair: a biomarker of targeted therapies (MAPK pathway)
Treatment should counteract pathway activation as a result of mutations in driving genes.
This type of effect is normally translated at the DNA Repair level.
By checking the DNA Repair signature before and after treatment, we can confirm the pathway's deactivation.
DNA Repair and response to immunotherapy
DNA Repair inhibitors
Synthetic lethality and cancer
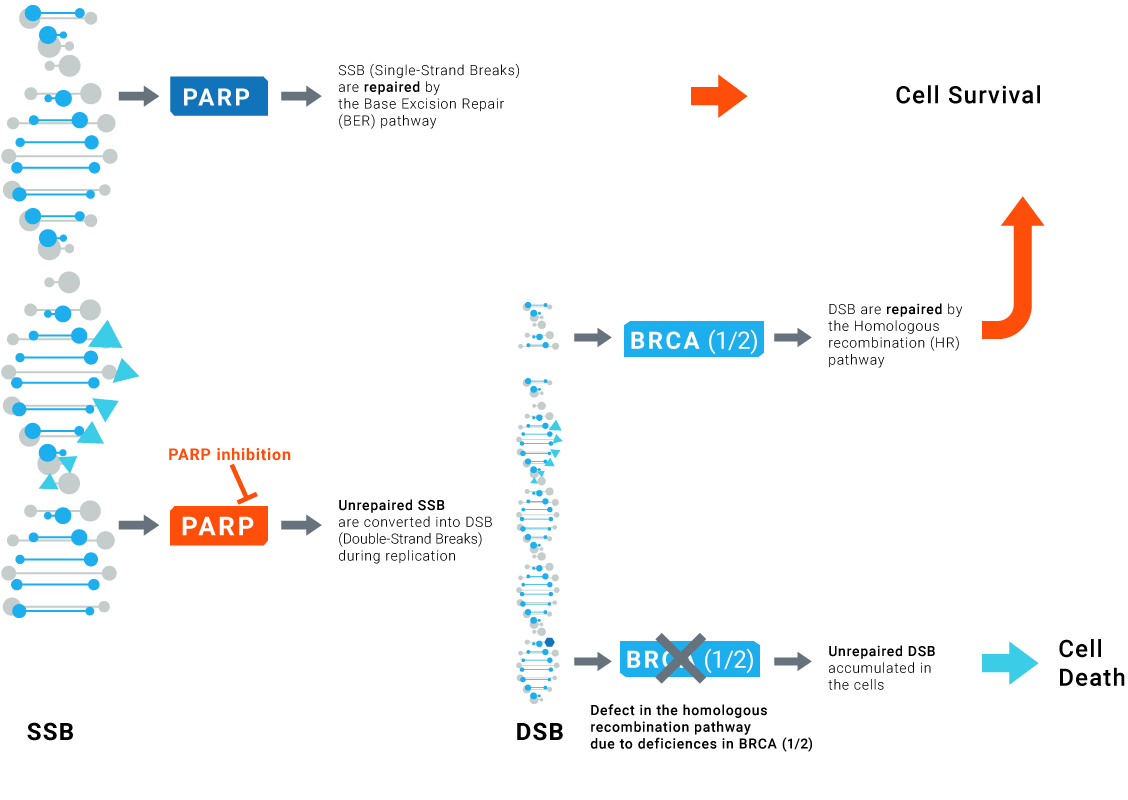
The concept of synthetic lethality is based on the inhibition of the alternative routes leading to the loss of function essential for cell survival and consequently to tumor cell death.
PARP inhibitors effectively kill tumors defective in homologous recombination pathway through the concept of synthetic lethality.
However, PARP inhibitors also show significant clinical benefit in patients without HR deficiencies.
Although some patients have sustained response to PARPi, resistance to PARPi arises in advanced disease.
LXRepair technologies can be used to check for the effective inhibition of specific DNA Repair pathways and can control the absence of any activity restoration potentially leading to resistance.
Particular case of DNA repair responses to PARP inhibition
Technology
SPOT-LX™ platform technology
Profiling DNA Repair
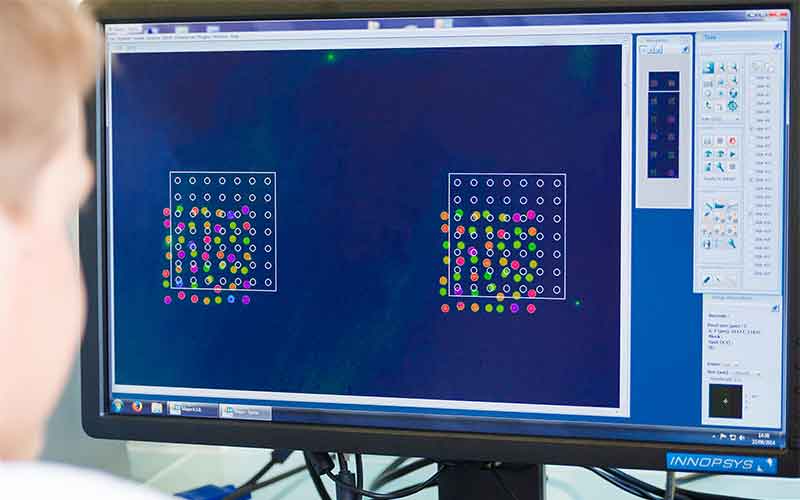
Multiple Advantages of our functional multiplex approach
- Reflects DNA Repair in real time
More relevant than genomic technologies - Characterizes the whole DNA Repair network
More powerful than single parameter assays - Integrates all molecular regulations in a single read-out
Our assays cover most DNA Repair pathways
3 complementary assays to cover most DNA Repair pathways and gain insights into major DNA Repair processes
- adapted to each therapeutic strategy
- designed with DNA Repair complexity in mind
- precise, complete and specific
Applications
LXRepair Assays and Clinical Diagnostics Applications
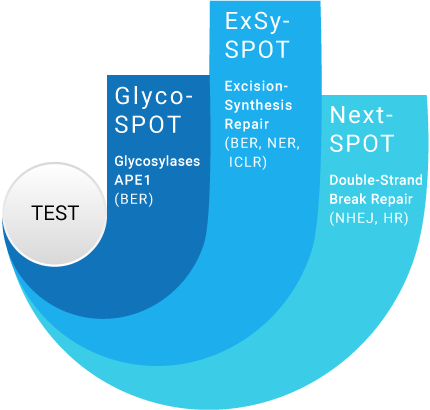
NER: Nucleotide Excision Repair
ICLR: Intra-Crosslink Repair
NHEJ: Non Homologous End Joining
HR: Homologous Recombination
Glyco-SPOT applications
Inflammation
DNA Repair inhibitor development
DNA Repair inhibitor resistance
ExSy-SPOT applications
DNA Repair inhibitor development
DNA Repair inhibitor resistance
Response to immunotherapy
Response to chemotherapy
Response to radiotherapy
Response to targeted therapy
Next-SPOT applications
DNA Repair inhibitor development
DNA Repair inhibitor resistance
DNA Repair
DNA Repair
Base Excision Repair
BER relies on Glycosylases to recognize and cleave "small" base lesions produced by alkylation, oxidation or deamination of normal DNA bases. The resulting abasic sites (AP sites) are then processed by an AP endonuclease. Bifunctional glycosylases display intrinsic AP lyase activity. The resulting gap is filled through the action of polymerases and finaly ligated.
Each glycosylase has a specific substrate spectrum.
LXRepair's Glyco-SPOT assay investigates repair enzymes by examining the substrates cleaved.
Nucleotide Excision Repair
Specific damage-sensing factors, including DDB1, DDB2, and XPC-hHR23B are involved in damage recognition (Global Genome Repair – GGR).
Depending on the structure of the lesion, different damage recognition factors and damage recognition factor enhancers are involved. Transcription-Coupled Repair (TCR) is initiated when RNA polymerase II stalls at a lesion. CSA and CSB are additional factors required for effective TCR. Transcription factor IIH (TFIIH) is recruited to the damaged site. XPA-RPA verifies whether the NER complex is correctly assembled, and helicases XPB and XPD unwind the double-helix. A short single-stranded DNA segment containing the lesion is then removed after incision by the endonucleases XPG and XPF-ERCC1. DNA polymerases fill the gap using the complementary sequence as a template. Finally, a DNA Ligase performs ligation.

Double-Strand Break Repair
DNA double-strand breaks (DSBs) are generated by ionizing radiations or endogenously during the replication process. They are among the most severe kinds of DNA damage.
DSBs are repaired through 2 main mechanisms that can operate in parallel or in competition, with various kinetics and outcomes: Homologous Recombination (HR) and Non-Homologous End Joining (NHEJ). NHEJ which is active through the whole cell cycle, is the predominant repair pathway. Alternative pathways such as Single-Strand Annealing (SSA) and alternative end joining pathways are also described. HR requires long homologous sequences and is predominantly error-free, while NHEJ tends to operate faster in a template-independent manner, at the cost of sequence alterations. SSA requires homologies flanking the DSB for operation and is a highly mutagenic process generating large deletions.